How to install r packages r studio
How to install r packages r studio
How do I install an R package from source?
A friend sent me along this great tutorial on webscraping The New York Times with R. I would really love to try it. However, the first step is to install a package called RJSONIO from source.
I know R reasonably well, but I have no idea how to install a package from source.
I’m running Mac OS X.
6 Answers 6
Trending sort
Trending sort is based off of the default sorting method — by highest score — but it boosts votes that have happened recently, helping to surface more up-to-date answers.
It falls back to sorting by highest score if no posts are trending.
Switch to Trending sort
If you have the file locally, then use install.packages() and set the repos=NULL :
Where path_to_file would represent the full path and file name:
Download the source package, open Terminal.app, navigate to the directory where you currently have the file, and then execute:
Do note that this will only succeed when either: a) the package does not need compilation or b) the needed system tools for compilation are present. See: R for Mac OS X
Madjoro$ R CMD INSTALL RJSONIO_0.2-3.tar.gz Warning: invalid package ‘RJSONIO_0.2-3.tar.gz’ Error: ERROR: no packages specified
You can install directly from the repository (note the type=»source» ):
A supplementarily handy (but trivial) tip for installing older version of packages from source.
First, if you call «install.packages», it always installs the latest package from repo. If you want to install the older version of packages, say for compatibility, you can call install.packages(«url_to_source», repo=NULL, type=»source»). For example:
Without manually downloading packages to the local disk and switching to the command line or installing from local disk, I found it is very convenient and simplify the call (one-step).
Plus: you can use this trick with devtools library’s dev_mode, in order to manage different versions of packages:
Earth Data Analytics Online Certificate
Earth analytics
earth-analytics Home
Lesson 3. Install & Use Packages in R
Install a Package
At the end of this activity, you will:
What is a Package?
A package, in R is a bundle of pre-built functionality. Think of it like a toolbox. Except for the tools may do things like calculate a mathematical function e.g. sum or create a plot.
Install a Package
Data tip You can install as many packages as you want in one string of code as follows install.packages(c(«name-one», «name-two»))
Call Package in R
Once the package is installed, to use it, you call the package at the top of your script like this:
Note that you don’t need to use quotes around the package name when you call it using the library() function. But you do need the quotes when you install a package.
In our case, the knitr and R Markdown packages load buttons and options within the R Studio environment that we can use. Thus we won’t have to call these two packages in our code in this lesson. However, when we use ggplot2 to plot, we will have to call it. We will see how calling a package works in a later set of lessons.
What Does Loading a Library Do?
Where Do R Packages Live?
R packages are most often found on the CRAN repository. When you call install.packages(«package-name-here») you are actually downloading the packages from CRAN. However there are other places where you may install packages from including:
Data tip While some R packages are just fine to use. Keep in mind that not all R packages are secure. Learn more.
You can also install multiple R packages at the same time. To do that, you send the install.packages() function a vector or list of package names.
A vector is a list of objects. the syntax for a vector is:
you can also store numbers in a vector.
Installing and Using R Packages
In our previous articles, we published i) guides for installing and launching R/RStudio, ii) the basics of R programming, and ii) guides for finding help in R.
Here, we’ll describe:
What is R packages?
An R package is an extension of R containing data sets and specific functions to solve specific questions.
R comes with standard (or base) packages, which contain the basic functions and data sets as well as standard statistical and graphical functions that allow R to work.
There are also thousands other R packages available for download and installation from CRAN, Bioconductor and GitHub repositories.
After installation, you must first load the package for using the functions in the package.
Installing R packages
Packages can be installed either from CRAN (for general packages), from Bioconductor (for biology-related packages) or from Github (developing versions of packages).
Install a package from CRAN
The function install.packages() is used to install a package from CRAN. The syntax is as follow:
For example, to install the package named readr, type this:
Note that, every time you install an R package, R may ask you to specify a CRAN mirror (or server). Choose one that’s close to your location, and R will connect to that server to download and install the package files.
It’s also possible to install multiple packages at the same time, as follow:
Install a package from Bioconductor
Bioconductor contains packages for analyzing biological related data. In the following R code, we want to install the R/Bioconductor package limma, which is dedicated to analyse genomic data.
To install a package from Bioconductor, use this:
Install a package from Github
GitHub is a repository useful for all software development and data analysis, including R packages. It makes sharing your package easy. You can read more about GitHub here: Git and GitHub, by Hadley Wickham.
To install a package from GitHub, the R package devtools (by Hadley Wickham) can be used. You should first install devtools if you don’t have it installed on your computer.
For example, the following R code installs the latest version of survminer R package developed by A. Kassambara (https://github.com/kassambara/survminer).
View the list of installed packages
To view the list of the already installed packages on your computer, type :
Note that, in RStudio, the list of installed packages are available in the lower right window under Packages tab (see the image below).
Folder containing installed packages
R packages are installed in a directory called library. The R function .libPaths() can be used to get the path to the library.
Load and use an R package
To use a specific function available in an R package, you have to load the R package using the function library().
In the following R code, we want to import a file (“http://www.sthda.com/upload/decathlon.txt”) into R using the R package readr, which has been installed in the previous section.
View loaded R packages
To view the list of loaded (or attached) packages during an R session, use the function search():
If you’re done with the package readr and you want to unload it, use the function detach():
Remove installed packages
To remove an installed R package, use the function remove.packages() as follow:
Update installed packages
If you want to update all installed R packages, type this:
To update specific installed packages, say readr and ggplot2, use this:
Summary
install.packages(“package_name”): Install a package
library(“package_name”): Load and use a package
detach(“package_name”, unload = TRUE): Unload a package
remove.packages(“package_name”): Remove an installed package from your computer
Related articles
Infos
This analysis has been performed using R software (ver. 3.2.3).
Enjoyed this article? I’d be very grateful if you’d help it spread by emailing it to a friend, or sharing it on Twitter, Facebook or Linked In.
Show me some love with the like buttons below. Thank you and please don’t forget to share and comment below!!
Avez vous aimé cet article? Je vous serais très reconnaissant si vous aidiez à sa diffusion en l’envoyant par courriel à un ami ou en le partageant sur Twitter, Facebook ou Linked In.
Recommended for You!
Recommended for you
This section contains best data science and self-development resources to help you on your path.
Data science
Popular Courses Launched in 2020
Trending Courses
Our Books
Others
Want to Learn More on R Programming and Data Science?
How to install packages
in R?
An R package is a library of functions that have been developed to cover some needs or specific scientific methods that are not implemented in base R. The functions that R provides by default are limited, so you might be wondering how to install new packages in R. In this tutorial we will review all the sources available to install R packages.
Installing R packages from CRAN
The Comprehensive R Archive Network (CRAN) is the official R packages repository, with thousands of free R packages available. Most of them have been developed by Data Scientists, Statisticians, Professors and researchers. There are all type of packages, from graphics packages as the well-known ggplot2 to very specific topics like the DTDA.cif package, that implements estimators for cumulative incidences of competing risks under double-truncation.
CRAN is the official R repository. All packages have been tested automatically and meet the CRAN policy.
First, you need to look for the name of the package you want to install. You may want to research for your topic googling something like: ‘graphics package R’ or ‘R package for time series’. You can also use the CRAN Task Views, where you can find the most relevant R packages by topic.
Take care! Just because a package is in CRAN does not imply that the methods are well implemented, since there are no third-party checks on this. Also, there may be errors that have not been checked. If you find any type of error or problem, contact the maintainer of the package.
Function to install R packages
Once you decided what package to install, just call the install.packages function with the name of the package inside the parenthesis with quotation marks. As an example, we are going to install the calendR package, that allows creating monthly and yearly calendars, but you can install the package you prefer.
Installing the CRAN packages with the menu
Alternatively, you can install R packages from the menu.
Installing packages in R from zip source
You can also set your working environment first with the setwd function to the folder where you have downloaded the package file and then install the package specifying the name of the zip or tar.gz file.
In case you have the zip hosted in some URL you can use the install.packages.zip function from the installr package. Note you can also install packages from CRAN (even older versions) this way.
Install multiple packages at once
If you need to install several packages at once without writing the same function over and over again, you can make use of the c function within the install.packages function. Note that now the quotation marks are needed to specify the packages names.
Now you know how to install R CRAN packages, but sometimes there are not all in CRAN for many reasons: CRAN has a code policy and some developers don’t want to spend time fixing minor issues to meet those requirements. Other times there exists a development version in GitHub of a CRAN package with additional features you may want. In the following sections you will learn how to install packages from other available sources.
In the following sections we will show you how to install unofficial source packages, but keep in mind that certain packages may be in a development process and could lead to unexpected errors sometimes.
Install R packages from GitHub or GitLab
GitHub is a well-known code sharing platform. If you go to the page, you can search for R packages using the search bar and writing something like: plot package language:R in case you want to look for graphics packages. Note that “language: R” is a search command of the page to restrict the results to only R code repositories. Suppose, for instance, that you want to download the development version of the ggplot2 package from GitHub. The URL would look like:
The first step is to install and load the devtools package, available in CRAN. In case you encounter some error means you also need to install the RTools.
Then you can call the install_github function with «account_name/repository_name» as argument to install the R package from GitHub.
Install R packages from R-Forge
Install bioconductor packages in R
Bioconductor is another project that hosts tools and R packages for analyzing biological data. First, you need to install the BiocManager package.
Second, you can make use of the install function of the package.
Note you can also install more than one package at the same time,
Install R package in Jupyter Notebook
If you are using R under the conda environment with Jupyter Notebook and you need more packages that the included like ‘Essentials’, you need to specify the repos argument as follows:
Update R packages
Updating R packages can be tedious if you have to reinstall the packages over and over again when some has a newer version. You can see the full list of your R packages that are not up-to-date with the old.packages function.
You will only be able to update a package if it is not loaded. If you have already loaded a package and you want to update it, use the detach function as follows: detach(package:some_package, unload = TRUE)
List functions in R package
You can also use the lsf.str or ls commands to list all the functions inside an attached (loaded) package.
Another option is to write: package_name:: and a list will show up in RStudio as a dropdown. In classic R you will have to press the tab button to show the functions on the screen, although it should be noted that if the package contains many functions not all will be shown, as is the case with the ggplot2 package:
View the source code of R package functions
Sometimes it can be interesting to inspect the code of any function. For that purpose, you have several options:
Note that, if the function is written in Fortan, C or any different language than R, you won’t be able to see the code with the first and the second method.
Check for installed packages
Sometimes you don’t remember if you have a package installed and you don’t want to waste your time reinstalling it. In order to avoid this, you can use the require function. Note that the main difference between require and library is that the first one returns a boolean and the second one returns an error if the package is not installed. The require function is designed to be used inside other functions.
The following line of code will also return TRUE if the package is installed, or FALSE if not.
Error: Cannot remove prior installation of package
If you encountered this error, you might be using different versions of R in the same computer. The solutions are:
Are you unable to install packages in R?
If you can’t install any package, there are many possible reasons:
If nothing works, try to close and open R again or try in another computer to verify if the problem persists.
A Installing R and RStudio
To get started with R, you need to acquire your own copy. This appendix will show you how to download R as well as RStudio, a software application that makes R easier to use. You’ll go from downloading R to opening your first R session.
Both R and RStudio are free and easy to download.
A.1 How to Download and Install R
R is maintained by an international team of developers who make the language available through the web page of The Comprehensive R Archive Network. The top of the web page provides three links for downloading R. Follow the link that describes your operating system: Windows, Mac, or Linux.
A.1.1 Windows
To install R on Windows, click the “Download R for Windows” link. Then click the “base” link. Next, click the first link at the top of the new page. This link should say something like “Download R 3.0.3 for Windows,” except the 3.0.3 will be replaced by the most current version of R. The link downloads an installer program, which installs the most up-to-date version of R for Windows. Run this program and step through the installation wizard that appears. The wizard will install R into your program files folders and place a shortcut in your Start menu. Note that you’ll need to have all of the appropriate administration privileges to install new software on your machine.
A.1.2 Mac
To install R on a Mac, click the “Download R for Mac” link. Next, click on the R-3.0.3 package link (or the package link for the most current release of R). An installer will download to guide you through the installation process, which is very easy. The installer lets you customize your installation, but the defaults will be suitable for most users. I’ve never found a reason to change them. If your computer requires a password before installing new progams, you’ll need it here.
Binaries Versus Source
R can be installed from precompiled binaries or built from source on any operating system. For Windows and Mac machines, installing R from binaries is extremely easy. The binary comes preloaded in its own installer. Although you can build R from source on these platforms, the process is much more complicated and won’t provide much benefit for most users. For Linux systems, the opposite is true. Precompiled binaries can be found for some systems, but it is much more common to build R from source files when installing on Linux. The download pages on CRAN’s website provide information about building R from source for the Windows, Mac, and Linux platforms.
A.1.3 Linux
R comes preinstalled on many Linux systems, but you’ll want the newest version of R if yours is out of date. The CRAN website provides files to build R from source on Debian, Redhat, SUSE, and Ubuntu systems under the link “Download R for Linux.” Click the link and then follow the directory trail to the version of Linux you wish to install on. The exact installation procedure will vary depending on the Linux system you use. CRAN guides the process by grouping each set of source files with documentation or README files that explain how to install on your system.
32-bit Versus 64-bit
R comes in both 32-bit and 64-bit versions. Which should you use? In most cases, it won’t matter. Both versions use 32-bit integers, which means they compute numbers to the same numerical precision. The difference occurs in the way each version manages memory. 64-bit R uses 64-bit memory pointers, and 32-bit R uses 32-bit memory pointers. This means 64-bit R has a larger memory space to use (and search through).
As a rule of thumb, 32-bit builds of R are faster than 64-bit builds, though not always. On the other hand, 64-bit builds can handle larger files and data sets with fewer memory management problems. In either version, the maximum allowable vector size tops out at around 2 billion elements. If your operating system doesn’t support 64-bit programs, or your RAM is less than 4 GB, 32-bit R is for you. The Windows and Mac installers will automatically install both versions if your system supports 64-bit R.
A.2 Using R
R isn’t a program that you can open and start using, like Microsoft Word or Internet Explorer. Instead, R is a computer language, like C, C++, or UNIX. You use R by writing commands in the R language and asking your computer to interpret them. In the old days, people ran R code in a UNIX terminal window—as if they were hackers in a movie from the 1980s. Now almost everyone uses R with an application called RStudio, and I recommend that you do, too.
R and UNIX
You can still run R in a UNIX or BASH window by typing the command:
which opens an R interpreter. You can then do your work and close the interpreter by running q() when you are finished.
A.3 RStudio
RStudio is an application like Microsoft Word—except that instead of helping you write in English, RStudio helps you write in R. I use RStudio throughout the book because it makes using R much easier. Also, the RStudio interface looks the same for Windows, Mac OS, and Linux. That will help me match the book to your personal experience.
You can download RStudio for free. Just click the “Download RStudio” button and follow the simple instructions that follow. Once you’ve installed RStudio, you can open it like any other program on your computer—usually by clicking an icon on your desktop.
The R GUIs
Windows and Mac users usually do not program from a terminal window, so the Windows and Mac downloads for R come with a simple program that opens a terminal-like window for you to run R code in. This is what opens when you click the R icon on your Windows or Mac computer. These programs do a little more than the basic terminal window, but not much. You may hear people refer to them as the Windows or Mac R GUIs.
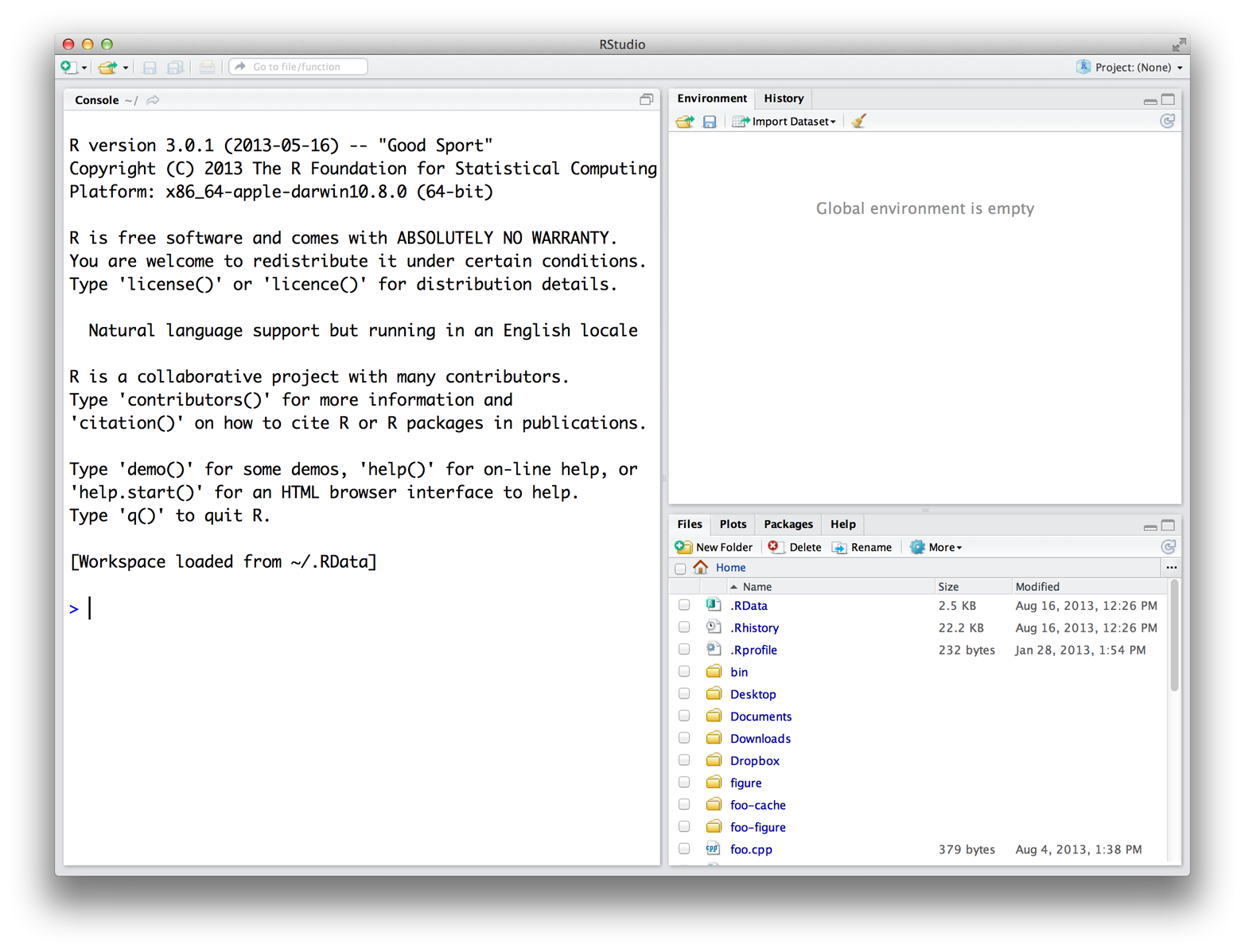
When you open RStudio, a window appears with three panes in it, as in Figure A.1. The largest pane is a console window. This is where you’ll run your R code and see results. The console window is exactly what you’d see if you ran R from a UNIX console or the Windows or Mac GUIs. Everything else you see is unique to RStudio. Hidden in the other panes are a text editor, a graphics window, a debugger, a file manager, and much more. You’ll learn about these panes as they become useful throughout the course of this book.
Figure A.1: The RStudio IDE for R.
Do I still need to download R?
Even if you use RStudio, you’ll still need to download R to your computer. RStudio helps you use the version of R that lives on your computer, but it doesn’t come with a version of R on its own.
A.4 Opening R
Now that you have both R and RStudio on your computer, you can begin using R by opening the RStudio program. Open RStudio just as you would any program, by clicking on its icon or by typing “RStudio” at the Windows Run prompt.